Environmental transmission
Accounting for space and uncertainty in real-time-location-system-derived contact networks
Information about the nature and quantity of contacts between organisms and their environment is critical for a variety of fields of study, including social behavior, habitat and resource use, and the epidemiology of infectious diseases. For instance, the frequency and duration of animals contacting each other may be indicative of their social relationship, or relative place within a dominance hierarchy. The paths that animals take through their environment may be indicative of their perception of the resources and risks around them. And the timing and proximity at which animals contact each other or parts of their environment can significantly influence the rate at which infectious pathogens get transmitted through a population. Thus, much may be gained from measuring the contacts of organisms with each other and the world in which they live.
The potential for scientific insight using contact analyses has greatly increased in recent years due to the development of technologies allowing for contact data to be collected at very high temporal and spatial resolutions. With these real-time location systems (RTLSs), individuals are outfitted with tracking devices (e.g., cell phones, GPS collars, RFID tags, etc.) and tracked continuously. While such data allow for unique research opportunities, they also present a number of conceptual and computational challenges. Below we describe some considerations of processing location data for contact analysis. We also present an innovative methodology for inferring the space and directionality of organisms to use in contact analysis, made available through a new R package called contact.
Contact Data Processing Considerations and Methods
Point-based data
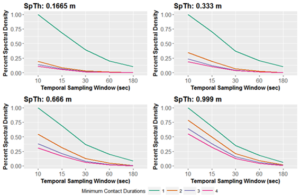
Most location data collected are ultimately point-based. That is, at each time point, tracked individuals’ locations are represented as a single geographic or planar coordinate pair (e.g., longitude/latitude, or x/y, respectively). Inter-point contact is generally determined by temporally aligning location data, then assessing spatial proximity of points at a particular spatial threshold of contact (SpTh) during a temporal sampling window (e.g., 10 seconds), for an assigned minimum duration necessary to ascribe a contact (MCD). In Dawson et al. (2019), we demonstrated that how point data is processed significantly influences the contact structure of the resulting dataset, and any inferences drawn from observed contacts. In particular, decisions about the SpTh, TSW, MCD, and the temporal aggregation of location datasets significantly influence derived contact data (Figure 1). One way to conceptualize this influence is by calculating the information content of contact datasets as Entropy, which measures the information loss that occurs as the criteria for counting something as a contact changes. We show that SpTh, TSW, and MCD interact to influence information content (i.e. entropy) of the contact structure of dataset by reducing information as criteria become more stringent (Figure 2). Furthermore, we show that these effects translate to epidemiological effects when contact data are used in infectious disease transmission models (Figure 3).
Polygon-based data
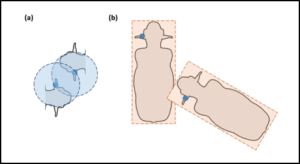
A particularly-interesting finding presented in Dawson et al. (2019)is that, when deriving contacts from point-based RTLS data using the methodology described above, information-content degrades with increasing SpTh values (Dawson et al. 2019). This suggests that proximity-based methods are insufficient for capturing and characterizing animal interactions involving body parts relatively far from tracking devices. This weakness, however, can be overcome by using polygons, rather than points, to represent tracked individuals and defining “contact” as instances of observed polygon intersections (Figure 4).
Uncertainty related to contact precision within relatively-large SpThs leads to epidemiologically- and sociologically-relevant interactions involving body parts not equipped with tags being potentially excluded from contact networks or misidentified as noise. Without this information, network modelers may draw incorrect conclusions regarding the frequency of inter-animal interactions, and pathogen-transmission potential in animal populations. In Farthing et al. (2019), we present novel procedures for deriving polygons from RTLS point data while maintaining distances and orientations associated with individuals’ relocation events. Furthermore, we demonstrate how highly versatile this methodology is for contact-network creation by generating inter-animal, environmental-, and visual-contact networks as examples. Ultimately, the methods described in Farthing et al. (2019) improve network-model realism and researchers’ ability to draw inferences from RTLS data, and can be used to answer a wide variety of epidemiological, ethological, and sociological research questions.
Contact R Package
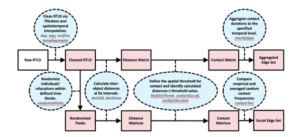
Our package, contact, contains 20+ functions in a processing pipeline to help researchers use RTLS data for contact and social network analyses (Figure 5). To use this package, users need only have a dataset containing point (i.e., latitude/longitude, or planar xy coordinates) or polygon (i.e., polygon shapefile) spatial data, individual IDs relating spatial data to specific individuals, and date/time information relating spatial data to temporal locations. Through package functions, users can create contact networks based on proximity- or intersection-based contact definitions, and can easily implement the novel methodologies described in Farthing et al. (2019). Furthermore, contact functions are not limited to describing inter-animal contacts, as users can also can quantify and characterize “contacts” between individuals and fixed areas (e.g., home ranges, waterbodies, buildings, etc.). As such, this package is an incredibly useful resource for facilitating epidemiological, ecological, ethological and sociological research.
This package can be found on the Lanzas Lab github page – https://github.com/lanzaslab/contact.
References
Dawson, D.E., Farthing, T.S., Sanderson, M.W., & Lanzas, C. 2019. Transmission on empirical dynamic contact networks is influenced by data processing decisions. Epidemics, 26: 32-42. https://doi.org/10.1016/j.epidem.2018.08.003.
Farthing, T.S., Dawson, D.E., Sanderson, M.W., & Lanzas, C. Accounting for space and uncertainty in real-time-location-system-derived contact networks. Ecology and Evolution, 10:4702-4715
Figure 1. Implications and visual explanations of processing criteria for dynamic contact data in disease transmission models.
Figure 2. The percent of maximum Entropy (information content; shown here as Fourier transformed spectral density) as processing criteria change for what constitutes a contact in a cattle feedlot contact dataset. As Spatial thresholds (SpTh) gets smaller, and temporal sampling windows and minimum contact (MCD) durations get larger, Entropy decreases.
Figure 3. The basic reproduction number (R0) of simulated epidemics increases as the Entropy of a contact dataset (shown here as log spectral density) increases due to processing decisions. SpTh value is indicated by color (orange = 0.1665 m, green = 0.333 m, blue=0.666 m, purple = 0.999 m); TSW is indicated by shape (circle = 10 sec; triangle = 15 sec; square = 30 sec; cross = 60 sec; x-box = 180 sec); MCD value is indicated by size.
Figure 4. Point-based vs. polygon-based contact definitions. (a) Contacts occur when points are observed within a predefined radial distance of one another. (b) Contacts occur when polygons intersect.
Figure 5. Pipeline to create time-aggregated contact and social networks using the contact package. Blue ovals describe necessary actions and relevant package functions. Red rectangles indicate function output.